Data Standardisation Working Group
Overview
The Data Standardisation Working Group of the International Bio-logging Society aims to fulfill the Society’s objective to “progress standardisation of data protocols used within the bio-logging community, with a view to making databases interoperable”. These standards should support the collection and use of bio-logging data to support research and applied uses. We welcome participation from all sectors, disciplines, career stages and regions.
Activities
Through community consultation during 2017–2024, we have identified a set of key needs for bio-logging data standards, illustrated below (from Davidson, Cagnacci et al., 2025). Members of this working group have previously helped to propose and demonstrate several potential standards, including De Pooter et al. (2017), Sequeira et al. (2021), van der Kolk et al. (2022), and MPIAB (2024). Moving forward, we acknowledge that the value of data standards relies on wide adoption of standards and ensuring accessibility of standardized data. Adoption will in turn depend on incentives to standardize, curate and publish data, on tools to steward long-term access to data that are not public, and on sustainable funding streams for these efforts. Our aim is for this working group to offer an international umbrella for coordination.
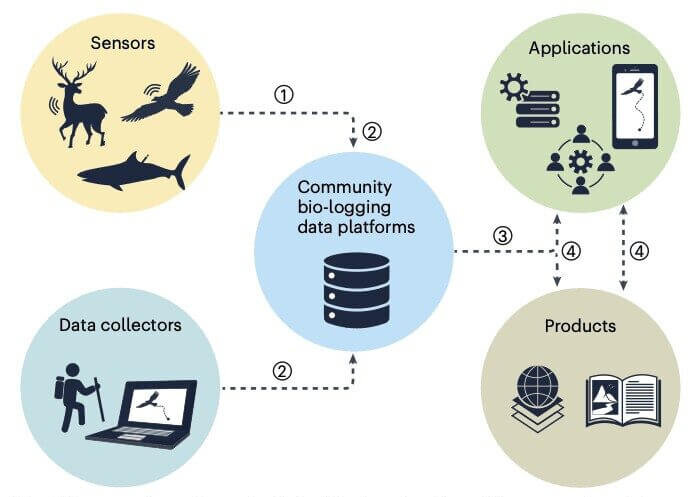
available for reuse (c). Pathways are not mutually exclusive, and datasets can proceed from a to c over time. Red dashed lines indicate eventual data loss if no additional steps are taken, and green ticks indicate long-term data preservation. Data standards would considerably enhance the discovery and reuse of data (from Davidson, Cagnacci et al., 2025).
Members
The current co-chairs of this working group are Sarah Davidson (Max Planck Institute of Animal Behavior, Germany), Peggy Newman (CSIRO, Australia), Francesca Cagnacci (Fondazione Edmund Mach, Italy). The advisory team includes Salma Abdel-Raheem, Max Czapanskiy, Luca Delucchi, Stacy DeRuiter, Connie Kot, Tina Odaka, Allison Payne, Katsufumi Sato, Ferdinando Urbano, and Qingshan Zhao.
We are building our advisory team of members to organize working group activities and communications. To get in touch, please contact ibls.datastandards AT gmail.com.
Resources
Visit the inventory of community initiatives.
Relevant outputs by working group members:
Davidson SC, Cagnacci F, Newman P, Dettki H, Urbano F, Desmet P, Bajona L, Bryant E, Carneiro APB, Dias MP, Fujioka E, Gambin D, Hunter C, Kato A, Kot CY, Kranstauber B, Lam CH, Lepage D, Naik H, Pye J, Sequeira AMM, Tsontos VM, van Loon E, Vo D, Rutz C. 2025. Establishing bio-logging data collections as living archives of animal life on Earth. Nature Ecol Evol. 9:204–213. https://doi.org/10.1038/s41559-024-02585-4
Desmet P. 2024. movepub: Prepare Movebank Data for Publication (Version 0.3.0) [Computer software]. https://github.com/inbo/movepub
Huybrechts P, Desmet P, Oldoni D, Van Hoey S. 2024. etn: Access Data from the European Tracking Network (Version 2.2.1) [Computer software]. https://github.com/inbo/etn
van der Kolk H-J, Desmet P, Oosterbeek K, Allen AM, Baptist MJ, Bom RA, Davidson SC, de Jong J, de Kroon H, Dijkstra B, et al. 2022. GPS tracking data of Eurasian oystercatchers (Haematopus ostralegus) from the Netherlands and Belgium. ZooKeys. 1123:31–45. https://doi.org/10.3897/zookeys.1123.90623
Sequeira AMM, O’Toole M, Keates TR, McDonnell LH, Braun CD, Hoenner X, Jaine FRA, Jonsen ID, Newman P, Pye J, et al. 2021. A standardisation framework for bio‐logging data to advance ecological research and conservation. Methods Ecol Evol. 12(6):996–1007. https://doi.org/10.1111/2041-210X.13593
Urbano, F., Cagnacci, F., & Euromammals Collaborative Initiative. 2021. Data management and sharing for collaborative science: Lessons learnt from the Euromammals initiative. Frontiers in Ecology and Evolution, 9, 727023. https://doi.org/10.3389/fevo.2021.727023
Czapanskiy M, Payne A, Hale C, Roche D, Nazario E, Kendall-Bar J, Buston R, Davidson SC, Clay T, Nisi A, et al. In preparation. Trends, challenges, and opportunities in open biologging data. https://flukeandfeather.github.io/openbiologging
Payne AR, Hale CM, Kendall-Bar J, Davidson SC, Beltran RS., Submitted. Minimum reporting standards can promote animal welfare and data quality in biologging research. Preprint at https://doi.org/10.32942/X29K7X
Sato, K., Watanabe, S., Noda, T., Koizumi, T., Yoda, K., Watanabe, Y.Y., Sakamoto, K.Q., Isokawa, T., Yoshida, M.A., Aoki, K., Takahashi, A., Iwata, T., Nishizawa, H., Maekawa, T., Kawabe, R. & Watanuki, Y. (2025) Biologging intelligent Platform (BiP): An Integrated and Standardized Platform for Sharing, Visualizing, and Analyzing Biologging Data. Movement Ecology, 13:23. https://doi.org/10.1186/s40462-025-00551-8
Contribute
Interested in joining the group or receiving email updates? Contact us at ibls.datastandards AT gmail.com.
